If pathogens like E. coli O157:H7 or Salmonella had a motto for survival, it might be: “Find! Bind! Multiply!”
That pretty much sums up what these food-poisoning bacteria do in nature, moving through our environment to find a host they can bind to and use as a staging area for multiplying and spreading.
But ARS food-safety scientists in California are determined to find out how to stop these and other foodborne pathogenic bacteria in their tracks, before the microbes can make their way to leafy greens and other favorite salad ingredients like tomatoes and sprouts.
The research is needed to help prevent the pathogens from turning up in fresh produce that we typically eat uncooked. That’s according to Robert E. Mandrell, who leads the ARS Produce Safety and Microbiology Research Unit. His team is based at the agency’s Western Regional Research Center in Albany, California.
The team is pulling apart the lives of these microbes to uncover the secrets of their success. It’s a complex challenge, in part because the microbes seem to effortlessly switch from one persona to the next. They are perhaps best known as residents of the intestines of warm-blooded animals, including humans. For another role, the pathogens have somehow learned to find, bind, and multiply in the world of green plants.
Sometimes the pathogenic microbes need the help of other microbial species to make the jump from animal inhabitant to plant resident. Surprisingly little is known about these powerful partnerships, Mandrell says. That’s why such alliances among microbes are one of several specific aspects of the pathogens’ lifestyles that the Albany scientists are investigating. In all, knowledge gleaned from these and other laboratory, greenhouse, and outdoor studies should lead to new, effective, environmentally friendly ways to thwart the pathogens before they have a chance to make us ill.
|
A Pathogen Targets Youngest Leaves
Knowing pathogens’ preferences is essential to any well-planned counter-attack. So microbiologist Maria T. Brandl is scrutinizing the little-understood ability of E. coli O157:H7 and Salmonella enterica to contaminate the elongated, slightly sweet leaves of romaine lettuce. With a University of California-Berkeley colleague, Brandl has shown that, if given a choice, E. coli has a strong preference for the young, inner leaves. The researchers exposed romaine lettuce leaves to E. coli and found that the microbe multiplied about 10 times more on the young leaves than on the older, middle ones. One explanation: The young leaves are a better nutrition “buy” for E. coli. “These leaves exude about three times more nitrogen and about one-and-one-half times more carbon than do the middle leaves,” says Brandl.
Scientists have known for decades that plants exude compounds from their leaves and roots that bacteria and fungi can use as food. But the romaine lettuce study, published earlier this year in Applied and Environmental Microbiology, is the first to document the different exudate levels among leaves of the two age classes. It’s also the first to show that E. coli can do more than just bind to lettuce leaves: It can multiply and spread on them.
|
Adding nitrogen to the middle leaves boosted E. coli growth, Brandl found. “In view of the key role of nitrogen in helping E. coli multiply on young leaves,” she says, “a strategy that minimizes use of nitrogen fertilizer in romaine lettuce fields may be worth investigating.”
In other studies using romaine lettuce and the popular herb cilantro as models, Brandl documented the extent to which E. coli and Salmonella are aided by Erwinia chrysanthemi, an organism that causes fresh produce to rot.
“When compared to plant pathogens, E. coli and Salmonella are not as ‘fit’ on plants,” Brandl says. But the presence of the rot-producing microbe helped E. coli and Salmonella grow on lettuce and cilantro leaves.
“Soft rot promoted formation of large aggregates, called ‘biofilms,’ of E. coli and Salmonella and increased their numbers by up to 100-fold,” she notes.
The study uncovered new details about genes that the food-poisoning pathogens kick into action when teamed up with plant pathogens such as soft rot microbes.
Brandl, in collaboration with Albany microbiologist Craig Parker, used a technique known as “microarray analysis” to spy on the genes. “The assays showed that Salmonella cells—living in soft rot lesions on lettuce and cilantro—had turned on some of the exact same genes that Salmonella uses when it infects humans or colonizes the intestines of animals,” she says. Some of these activated genes were ones that Salmonella uses to get energy from several natural compounds common to both green plants and to the animal intestines that Salmonella calls home.
A One-Two Punch to Tomatoes
Salmonella also benefits from the presence of another plant pathogen, specifically, Xanthomonas campestris, the culprit in a disease known as “bacterial leaf spot of tomato.” But the relationship between Salmonella and X. campestris may be different than the relation of Salmonella to the soft rot pathogen. Notably, Salmonella benefits even if the bacterial spot pathogen is at very low levels—so low that the plant doesn’t have the disease or any visible symptoms of it.
That’s among the first-of-a-kind findings that microbiologist Jeri D. Barak found in her tests with tomato seeds exposed to the bacterial spot microbe and then planted in soil that had been irrigated with water contaminated with S. enterica.
In a recent article in PLoS ONE, Barak reported that S. enterica populations were significantly higher in tomato plants that had also been colonized by X. campestris. In some cases, Salmonella couldn’t bind to and grow on—or in—tomato plants without the presence of X. campestris, she found.
|
“We think that X. campestris may disable the plant immune response—a feat that allows both it and Salmonella to multiply,” she says.
The study was the first to report that even as long as 6 weeks after soil was flooded with Salmonella-contaminated water, the microbe was capable of binding to tomato seeds planted in the tainted soil and, later, of spreading to the plant.
“These results suggest that any contamination that introduces Salmonella from any source into the environment—whether that source is irrigation water, improperly composted manure, or even insects—could lead to subsequent crop contamination,” Barak says. “That’s true even if substantial time has passed since the soil was first contaminated.”
Crop debris can also serve as a reservoir of viable Salmonella for at least a week, Barak’s study showed. For her investigation, the debris was composed of mulched, Salmonella-contaminated tomato plants mixed with uncontaminated soil.
“Replanting fields shortly after harvesting the previous crop is a common practice in farming of lettuce and tomatoes,” she says. The schedule allows only a very short time for crop debris to decompose. “Our results suggest that fields known to have been contaminated with S. enterica could benefit from an extended fallow period, perhaps of at least a few weeks.”
Ordinary Microbe Foils E. coli
While the bacterial spot and soft rot microbes make life easier for certain foodborne pathogens, other microbes may make the pathogens’ existence more difficult. Geneticist Michael B. Cooley and microbiologist William G. Miller at Albany have shown the remarkable effects of one such microbe, Enterobacter asburiae. This common, farm-and-garden-friendly microorganism lives peaceably on beans, cotton, and cucumbers.
In one experiment, E. asburiae significantly reduced levels of E. coli and Salmonella when all three species of microbes were inoculated on seeds of thale cress, a small plant often chosen for laboratory tests.
The study, published in Applied and Environmental Microbiology in 2003, led to followup experiments with green leaf lettuce. In that battle of the microbes, another rather ordinary bacterium, Wausteria paucula, turned out to be E. coli’s new best friend, enhancing the pathogen’s survival sixfold on lettuce leaves.
“It was the first clear example of a microbe’s supporting a human pathogen on a plant,” notes Cooley, who documented the findings in the Journal of Food Protection in 2006.
But E. asburiae more than evened the score, decreasing E. coli survival 20- to 30-fold on lettuce leaves exposed to those two species of microbes.
The mechanisms underlying the competition between E. asburiae and E. coli are still a mystery, says Cooley, “especially the competition that takes place on leaves or other plant surfaces.”
Nevertheless, E. asburiae shows initial promise of becoming a notable biological control agent to protect fresh salad greens or other crops from pathogen invaders. With further work, the approach could become one of several science-based solutions that will help keep our salads safe.—By Marcia Wood, Agricultural Research Service Information Staff.
This research is part of Food Safety, an ARS national program (#108) described on the World Wide Web at www.nps.ars.usda.gov.
To reach scientists mentioned in this article, contact Marcia Wood, USDA-ARS Information Staff, 5601 Sunnyside Ave., Beltsville, MD 20705-5129; phone (301) 504-1662, fax (301) 504-1486.
|
What Genes Help Microbes Invade Leafy Greens?
When unwanted microbes form an attachment, the consequences—for us—can be serious.
That’s if the microbes happen to be human pathogens like Listeria monocytogenes or Salmonella enterica and if the target of their attentions happens to be fresh vegetables often served raw, such as cabbage or the sprouted seeds of alfalfa.
Scientists don’t yet fully understand how the malevolent microbes form colonies that cling stubbornly to and spread across plant surfaces, such as the bumpy leaves of a cabbage or the ultra-fine root hairs of a tender alfalfa sprout.
But food safety researchers at the ARS Western Regional Research Center in Albany, California, are putting together pieces of the pathogen puzzle.
A 1981 food-poisoning incident in Canada, caused by L. monocytogenes in coleslaw, led microbiologist Lisa A. Gorski to study the microbe’s interactions with cabbage. Gorski, with the center’s Produce Safety and Microbiology Research Unit, used advanced techniques not widely available at the time of the cabbage contamination.
“Very little is known about interactions between Listeria and plants,” says Gorski, whose study revealed the genes that Listeria uses during a successful cabbage-patch invasion.
The result was the first-ever documentation of Listeria genes in action on cabbage leaves. Gorski, along with coinvestigator Jeffrey D. Palumbo—now with the center’s Plant Mycotoxin Research Unit—and others, documented the investigation in a 2005 article in Applied and Environmental Microbiology.
Listeria, Behaving Badly
“People had looked at genes that Listeria turns on, or ‘expresses,’ when it’s grown on agar gel in a laboratory,” says Gorski. “But no one had looked at genes that Listeria expresses when it grows on a vegetable.
“We were surprised to find that when invading cabbage, Listeria calls into play some of the same genes routinely used by microbes that are conventionally associated with plants. Listeria is usually thought of as a pathogen of humans. We hadn’t really expected to see it behaving like a traditional, benign inhabitant of a green plant.
“It’s still a relatively new face for Listeria, and requires a whole new way of thinking about it.”
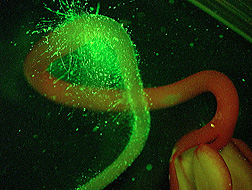
In related work, Gorski is homing in on genetic differences that may explain the widely varying ability of eight different Listeria strains to successfully colonize root hairs of alfalfa sprouts—and to resist being washed off by water.
In a 2004 article in the Journal of Food Protection, Gorski, Palumbo, and former Albany associate Kimanh D. Nguyen reported those differences. Poorly attaching strains formed fewer than 10 Listeria cells per sprout during the lab experiment, while the more adept colonizers formed more than 100,000 cells per sprout.
Salmonella’s Cling Genes
Colleague Jeri D. Barak, a microbiologist at Albany, led another sprout investigation, this time probing the ability of S. enterica to attach to alfalfa sprouts. From a pool of 6,000 genetically different Salmonella samples, Barak, Gorski, and coinvestigators found 20 that were unable to attach strongly to sprouts.
Scientists elsewhere had already identified some genes as necessary for Salmonella to successfully invade and attach to the guts of animals such as cows and chickens. In the Albany experiments, some of those same genes were disrupted in the Salmonella specimens that couldn’t cling to alfalfa sprouts.
Their 2005 article in Applied and Environmental Microbiology helped set the stage for followup studies to tease out other genes that Salmonella uses when it is living on and in plants.
A deeper understanding of those and other genes may lead to sophisticated defense strategies to protect tomorrow’s salad greens—and us.—By Marcia Wood, Agricultural Research Service Information Staff.
|
Environmental Surveillance Exposes a Killer
It started as a manhunt for a microbe, but it became one of the nation’s most intensive farmscape searches for the rogue pathogen E. coli O157:H7.
ARS microbiologist Robert E. Mandrell and geneticist Michael B. Cooley of the Produce Safety and Microbiology Research Unit in Albany, California, had already been collaborating in their own small-scale study of potential sources of E. coli O157:H7 in the state’s produce-rich Salinas Valley when, in 2005, they were asked to join another one. The new investigation became a 19-month surveillance—by the two scientists and other federal and state experts—of E. coli in Salinas Valley watersheds.
“It may seem like an obvious concept today,” says Mandrell, “but at the time, there was little proof that E. coli contamination of produce before harvest could be a major cause of food-poisoning outbreaks.”
Mandrell and Cooley aided the California Food Emergency Response Team, as this food-detective squad was named, in tracing movement of E. coli through the fertile valley. This surveillance showed that E. coli O157:H7 can travel long distances in streamwater and floodwater.
In 2006, E. coli O157:H7 strains indistinguishable from those causing human illness associated with baby spinach were discovered in environmental samples—including water—taken from a Salinas Valley ranch.
Wild pigs were added to the list of animal carriers of the pathogen when one of the so-called “outbreak strains” of E. coli O157:H7 was discovered in their dung. The team documented its work in 2007 in PLoS ONE and Emerging Infectious Diseases.
The Albany scientists used a relatively new technique to detect E. coli O157:H7 in water. Developed at the ARS Meat Animal Research Center in Clay Center, Nebraska, for animal hides, the method was adapted by the Albany team for the outdoor reconnaissance.
Because of their colleagues’ work, says Cooley, “We had the right method at the right time.”—By Marcia Wood, Agricultural Research Service Information Staff.
"Outmaneuvering Foodborne Pathogens" was published in the July 2008 issue of Agricultural Research magazine.
http://www.ars.usda.gov/is














No comments:
Post a Comment